CONSERVATION GENETICS
Conservation biology is a rapidly growing discipline of cology and evolutionary
biology. In many ways the issues surrounding the conservation of endangered
or threatened species have rejuvenated aspects of population genetics and
systematics that were often viewed as "academic." Indeed, may
aspects of conservation biology can be view as "applied" ecology
and evolutionary biology.
We will consider two different approaches to conservation genetics:
1) population genetic issues relating to the maintenance of genetic variability,
and 2) systematics issues relating to the description of biodiversity and
the recognition of evolutionary "units" for preservation.
Due to the rapid destruction of habitat there are many species that
are going extinct. One estimate is in the neighborhood of 100 species per
day! Habitat destruction is generally attributable to human impact, but
the causes of extinction are varied: environmental variability, natural
catastrophes, demographic variability (stochasticity), genetic stochasticity,
etc. Faced with this problem, biologist set out to determine a Minimum
Viable Population Size (MVP): a population size that ensure the persistence
of a species for specified period of time. One description is a 99% chance
of persistence of 1000 years.
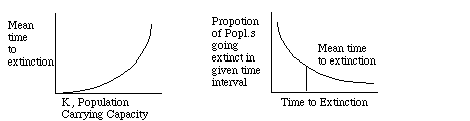
Theoretical population genetics simulations have lead to some predictions. There should be a positive relation between population carrying capacity (population size the (local) environment can sustain) and the average time to population extinction. Moreover, extinction times are exponentially distributed so a large proportion of populations will go extinct in a period of time less than the mean time to extinction.

The implication from these simulation results is that Larger Populations
are Better: it will take longer for a larger population to go extinct,
and larger populations will lower the extinction curve. If environmental
stochasticity is added to these models, theory suggests that MVP should
be 500 - 1000. If demographic stochasticity (randomly fluctuating birth
and death rates, i.e., random population growth rates), MVP needs to be
sustained at higher values (1000 - 5000). If this system is placed in the
context of an interwoven ecosystem, the MVP should probably be higher.
MVP and Genetic variation Population genetic theory indicates
that inbreeding depression will be likely with an effective population
size of Ne < 50. To avoid the possibility of inbreeding,
a lower limit of MVP = 500. These numbers may seem like obscure
conclusions from a series of complex Populus simulations, but they are
the working numbers for policy issues : N = 50 defines the critical
list; N = 500 defines the endangered list.
The values for MVP and "critical" versus "endangered"
lists stem from some basic issues relating to effective population size.
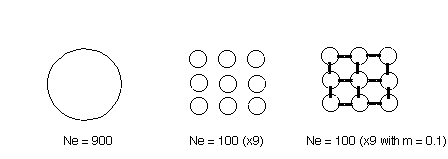
Ne refers to the effective number of breeders in the population,
and can be affected greatly by variance in reproductive success
A focus here is the ratio of effective size to census size Ne/N
ratio. Observations from field and laboratory experiments indicate
that Ne/N ratios are about 0.25 (range = 0.05 to 1.0). This
means that either some individuals are breeding and others are not, or
that some individuals have different degrees reproductive success
that others. It is possible that a substantial proportion of the population
does reproduce, but that a small number of individuals produce most of
offspring reducing the genetic pool from which alleles are drawn. This
reduces the Ne/N ratio and hence brings the population closer
to the demographic danger zone.
With a Ne/N ratio of 0.25, the census size should
be 4 times higher than the simple numbers predicted by "critical"
and "endangered" estimates. Now add population size fluctuations:
bottlenecks in census size affect Ne more severely. Recall:
= (1/Ne) = (1/t) (1/Ne)
The net effect of these factors is that MVP should be 5X to 10X Ne.
Given a finite amount of space for a nature preserve, do you establish
a Single Large or Several Small (SLOSS) system. The answer
depends on the likely causes of extinction in the particular system of
concern. If the species is subject to demographic fluctuations, it would
be better to maintain one large system, since the plot above suggests that
extinction is more likely in small populations. In systems of species where
environmental stochasticity is a general problem , the Several Small approach
is probably better: many sub-reserves will reduce the chances of losing
the entire system.
The SLOSS debate is closely related to the dynamics of Metapopulations.
The maintenance of genetic diversity can be enhanced by structuring in
a metapopulation system. Alleles that might be lost in one deme can be
fixed in another, and the average of the metapopulation system may
maintain more heterozygosity than a simple population of similar total
population size. The solution to this is not general since metapopulation
systems may have varying degrees of migration between demes (at some level
of migration, metapopulations become 'systems of subpopulations' since
in the strict sense the demes of a metapopulation experience little gene
flow.)
Metapopulations can also contribute to the purging of deleterious recessive alleles. With some level of inbreeding in demes, deleterious recessives will be selected against. With limited amounts of gene flow, the system can effectively purge these alleles that might not be expressed in a large random mating population. One approach is to have semi- isolated subpopulations with corridors for dispersal.
There is no one solution to all these problems. The answers depend on
1) the species and ecosystem in question, 2) the demographic issues (constant
or variable) and 3) the existing levels of genetic variation.
Many issues in conservation genetics have been centered around Zoo
Biology. Most zoos maintain rare or endangered species and are involved
in captive breeding programs with such species. Again, a central
issue is the maintenance of genetic variation. A number of recent studies
have addressed the captive breeding protocol to determine how mating
systems affect the maintenance of genetic variation.
Using Drosophila, several studies have shown that populations maintained
with equal founder size (EFS) retain more genetic variation
that populations maintained by random mating. EFS approaches equalize the
number of founders that contribute to the "captive" population
each generation. Similarly, equal founder representation (EFR) studies
retain slightly more genetic variation than randomly mating populations.
EFR populations are maintained with a controlled pedigree where the parentage
of each contributing female and male is known. These types of studies use
allozyme electrophoresis to study directly the levels of heterozygosity
in experimental (EFS, EFR) and randomly mating control populations over
time. In addition, fitness studies can be performed by competing
experimental flies against tester stocks to determine if a higher
fitness is maintained. The conclusion from these studies is that controlled
mating schemes can make a difference in retailing genetic variation
and attaining higher levels of fitness. The next step is how do you translate
these findings into captive breeding of Panda Bears??
Phylogenetic approaches to conservation biology have received
a lot of attention. This follows directly from the general concern about
Biodiversity. To properly appreciate and understand biodiversity,
we must have a sense of phylogenetic structure of the taxa involved. This
applies to broad levels of organization (soil bacteria, plants, animals)
as well as to smaller taxonomic units (populations within species). Molecular
systematic approaches have been of great use since many new techniques
can be applied without harming wild individuals, and can even be applied
to museum skins for historical comparisons. In the context of the Endangered
Species Act several important issues come up: What is the phylogenetic
relationship of the endangered species? How much and what type of
genetic variation (gene trees)? What do we preserve? What IS a species?
Any genetically distinct entity has evolutionary potential.
Two case studies: The Dusky seaside sparrow: The species declined
in the 1960's; by 1980 only 6 birds remained that were all male.
A captive breeding program was initiated and captive Scott's seaside
sparrow was chosen as the females. When the last male Dusky died Avise
& Nelson analyzed its mitochondrial DNA (mtDNA) and found that the
Dusky and Scott's seaside sparrows were members of different clades on
the phylogeny of these sparrows. The implication is that more detailed
phylogenetic knowledge of the endangered species would have lead to different
management decisions in handling this captive breeding program (choosing
a different species to mate to the Dusky)
The red wolf was placed into a captive breeding program in 1974.
By 1975 it was extinct in the wild. Early data suggested that red wolves
hybridized with coyotes. Since coyote populations do well in human-disturbed
habitats, hybridization may have affected the survival of the red wolf.
Wayne & Jenks studied the mitochondrial DNA from captive red wolves
and from 77 animals collected from the wild during the capture program.
They also used the polymerase chain reaction (PCR) to sequence mitochondrial
DNA from museum skins ("ancient DNA techniques") collected before
hybridization between red wolves and coyotes is thought to have begun.
They found that red wolves have either a gray wolf or a coyote mtDNA,
indicating that the red wolf "species" is entirely a hybrid.
Other researchers disagree about the species "status" of the
red wolf. Nevertheless, this raises the question: What should
we protect? If the species isn't really a clear entity phylogenetically,
does it deserve a conservation/captive breeding effort? Wayne & Jenks
argue that their data should not be used to advocate the discontinuation
of the conservation effort of the red wolf.
These examples illustrate why the recognition of Evolutionarily Significant
Units (ESU) is an issue of great concern in conservation genetics.
ESUs are defined various ways, but they are recognized as populations with
independent evolutionary histories. Fixed allelic differences or strong
phylogenetic support such as multiple synapomorphies distinguishing
one population from another are good grounds for the recognition of distinct
ESUs. Hence, a full understanding of how to do molecular systematics is
very important in molecular conservation genetics.
It should be emphasized that mitochondrial DNA markers are maternally
inherited and may not reflect the true evolutionary history of the entire
populations. Hence it is advisable to have additional nuclear markers for
ESU recognition such as allozymes, RAPDs or microsatellites. RAPDs
are Randomly Amplified Polymorphic DNA. This method uses the polymerase
chain reaction to amplify random regions of the genome with short random
10-base primers. Microsatellites are regions of the genome that vary in
the number of tandemly repeated sequences. The repeats are short (2-4 base
pair repeat unit), but there may be many of them in a row. Individuals
differ in the number of repeat units they have and this difference can
be determined by gel electrophoresis.
Another relevant level of concern is a Management Unit (MU).
These are defined as populations that have different frequencies of alleles,
but do not necessarily show fixed differences between populations.
Hence several MUs may exist within an ESU. The figures below illustrate
the difference. The general lesson is that molecular approaches to conservation
biology are potentially highly informative since many overt phenotypic
characteristics cannot reveal important differences that distinguish populations.
Since conserving endangered species is inherently a "genetic"
endeavor, to the extent that we recognize species as discrete reservoirs
of historically unique genetic material, the molecular approaches are very
useful.