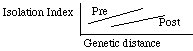
GENETICS OF SPECIATION
Speciation has occurred, is occurring and will occur. These are undeniable
facts. The problem is: How do we study speciation? There is no single
approach since there is no single mechanism by which species speciate.
Some approaches used in the past:
Study patterns of morphological change in the fossil record in
a well defined lineage of organisms. Success depends on many unknowns:
stratigraphic resolution (will you "see" the speciation
event); distinguishing geographic variants from true species (all
you have is morphology.
Comparisons of closely related species. These have speciated
recently (assuming closely related ~ short time since speciation) so careful
studies of their biology may identify important features that contribute
to reproductive isolation.
Study intraspecific variation. Look for evidence of incipient
barriers to gene exchange. Perform crosses between individuals from different
regions; look for differences in genital morphology, secondary sexual characteristics.
These may show some bimodal distribution suggestive of early steps in evolution.
Must ask: what might we expect to find? This depends entirely on the model
of speciation that might apply to the organism under study. Looking within
a large species range for signs of variation may be fruitless if the speciation
mode is peripatric with genetic revolutions?
Laboratory populations might serve as model systems. One can
establish the conditions of the specific model under question and ask if
the predicted divergence is observed. Mathematical models can address
specific predictions about modes of speciation. Both of these "artificial"
methods are important since they can identify what is possible.
Knowing what's possible might spur one on to looking for it in unexpected
contexts in natural populations.
With the use of molecular tools the comparisons of intraspecific and
interspecific genetic variation has been studied in some detail. Aim is
to identify genetic changes during speciation. These data show us
that genetic change is associated with speciation. We want to be able to
describe the genetics of speciation and the genetics of species
differences. To do so we need to distinguish genetic changes
that cause speciation from those that accompany speciation.
These will differ a lot from one group of organisms to the next and will
depend on the genetic architecture of speciation. Best data on both
of these issues have come from the many species of Drosophila
Coyne and Orr (1989, Evolution vol. 43, pg. 362-381) take Ayala's
approach one step further and attempt to correlate genetic distance (Nei's
D) with amounts of prezygotic and postzygotic isolation. In the literature
there are many reports of the amount of genetic distance between closely
related species of Drosophila and the amount of reproductive isolation
between many of the species for which genetic distance has been measured
(premating or prezygotic isolation is measured as [1-(proportion
of heterotypic matings/proportion of homotypic matings)] which ranges from
- infinity for all heterotypic (between species) matings to 0 for random
mating to +1 for all homotypic matings. Rarely do two species prefer to
mate with the wrong type so the index effectively ranges from 0 to 1).
Postzygotic or postmating isolation can be measured as in the
following example. Consider two species, A and B. These can be crossed
two ways (reciprocally) to produce hybrid offspring. We can also
examine the viability or fertility of the two sexes of these hybrid offspring,
hence four contexts are examined to score postzygotic isolation:
| Case | Female parent | Male parent | Offspring | Inviable or sterile? | |
| 1 | Species A | Species B | Male | No = 0 | Yes = 1 |
| 2 | Species A | Species B | Male | No = 0 | No = 0 |
| 3 | Species B | Species A | Female | No = 0 | No = 0 |
| 4 | Species B | Species A | Female | No = 0 | No = 0 |
| I = 0 | I = .25 |
Note: Isolation index is the average score for the four cases.
In any particular case one could choose to score isolation in terms
of the presence or absence of either isolation or sterility. Normally
hybrid sterility evolves before hybrid inviability (mules are sterile
but viable). Hence an index based on sterility would have higher values
than an index based only on evidence for inviable hybrid offspring.
Coyne and Orr extracted these two types of data from the literature
and tested some important ideas about the genetics of speciation. The general
idea is that genetic distance (D) is positively related to time
(the molecular clock hypothesis) and thus species pairs showing different
degrees of genetic distance should be at different degrees of completion
of the speciation process (be aware that many organisms are in the
process of speciating as you read these notes). Coyne and Orr show that
there is a significant relationship between genetic distance and both premating
and postmating isolation 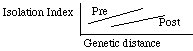
Two interesting additional points: sympatric species show greater
prezygotic isolation than allopatric species pairs. This pattern is
consistent with the reinforcement hypothesis and suggest that reinforcement
can act (see figs. 16.14 - 16.16, pg. 454-455). A second observation:
less genetic distance between species pairs that produce sterile
or inviable males than between species pairs that produce sterile
or inviable females (D(A-B)sterile males
< D(A-B)sterile females).
This observation confirmed a well documented pattern known as Haldane's
Rule (see table 15.2, pg. 406) stating that when hybrid crosses produce
sterile or inviable offspring, the sex that exhibits this is most likely
the heterogametic sex (the sex with two different sex chromosomes,
e.g. X and Y in male humans and Drosophila; in birds and butterflies
the female is heterogametic with Z and W). Another "rule"
of speciation is that genes affecting reproductive isolation are
typically found on the X chromosome (where X is the "female"
chromosome; see another paper by Coyne and Orr: "Two Rules of Speciation",
in Speciation and its Consequences, 1989, D. Otte & J. Endler,
editors, Sinauer Associates).
The current belief about the large "X effect" is that advantageous
mutations are more likely to accumulate on the X since it is hemizygous
in males, so half of the time recessive advantageous mutations will be
expressed. Similar mutations occurring on autosomes will be less likely
to be expressed because autosomes are always paired and an advantageous
mutation would have to be dominant to be "visible" to selection.
Thus diverging populations (incipient species) will tend to accumulate
different mutations on their respective X chromosomes. When individuals
are crossed between these divergent populations, there will deleterious
pleitropic interaction effects between these new alleles on the X and other
genes throughout the genome. The new mutations certainly were not deleterious
when they arose within each separated population, but when paired with
autosomes from a diverged population these mutations do not function properly,
thus one would only see the effect in a hybrid cross. See table
16.2, pg. 456.
Attempts to identify genes that keep species isolated go back to Dobzhansky
in the 1930's: crosses between D. pseudoobscura and D. persimilis
produce sterile males and fertile females as F1 hybrids. These F1 females
can be backcrossed to males of either species, so the backcrossed offspring
can have all combinations of chromosomes. With four chromosome pairs in
each species, the F1 hybrid will have four heterokaryotypic pairs
of chromosomes. The two possible backcrosses (one in each direction)
can result in 16 possible combinations of chromosomes. Frequently
find that the offspring with nonmotile sperm (= sterile) are the ones with
sex chromosomes from each species (see figures). Deleterious interactions
between sex chromosomes and/or between sex chromosomes and autosomes are
implied, but the details are the topic of a lot of current research (see
Orr 1993, Nature vol. 361, pg. 532 & pg. 496). These types of
experiments, coupled with molecular biology may someday allow us to identify
the genes and the types of changes that can lead to speciation. Again,
we would like to know the genetic architecture of speciation: how
many genes involved?; what sorts of mutations at each gene? what sorts
of interactions among genes? etc.