MODELS OF SPECIATION
Speciation is a fundamental issue in evolutionary biology, but it is
both fascinating and frustrating: we know it does happen but it
its an historical phenomenon so it is difficult to observe. The
two camps of evolutionary biologists best equipped to deal with speciation
(in terms of mechanism, population geneticists; in terms of time-frames,
paleontologists) are both incapable of "seeing" speciation except
in very special situations. We must rely on strong inference to
properly understand speciation. This inference is in many cases very rigorous
and scientific although it is historical, i.e., requires an interpretation
of what has gone on in the past.
Defining speciation depends on one's species concept. (Recall species concepts: typological, evolutionary, biological, recognition). In its simplest form speciation is lineage splitting; the resulting lineages are genetically isolated and ecologically distinct. This implies that something intrinsic about the new lineages (an aspect of its biology, e.g., genetics) makes/keeps them distinct. Speciation then must involve the evolution of intrinsic barriers to gene exchange. Intrinsic barriers can related in many ways to extrinsic barriers to gene exchange (abiotic factors limiting gene flow: rivers, isolated islands, glaciers). A variant of a species could be adapted to live in a particular environment that is spatially distinct from other types of environmental conditions; here an intrinsic component contributes to an extrinsic barrier. The notion of the evolution of barriers to gene exchange applies to virtually all species concepts since unlimited gene exchange between two populations/species would prevent the evolution of the defining principles of a given species concept: 1) true typological differences must have a genetic basis, 2) evolutionary lineages would not have their own "evolutionary tendencies" with homogenization due to gene exchange, 3) reproductive isolation (either pre- or postmating) would not be maintained with unlimited gene flow, and 4) mate recognition systems could not be maintained as distinct with unlimited gene exchange.
Without the evolution of some intrinsic barrier to gene exchange,
fusion of the two incipient species would be one likely outcome
(populations would blend back into one), or extinction of one or
the other lineages (one population out competed [at the individual level!]
the diverged sister population leaving only one population).
MODELS OF SPECIATION
There are may models which have been proposed that enable barriers to
gene exchange to evolve; as argued by Ernst Mayr, geographic isolation
provides the most effective barrier. We thus consider the allopatric
model:
1. continuous distribution split into two (or more) sub populations
2. differentiation in allopatry (different selection regimes; not necessarily selection for speciation)
3. if populations come into secondary contact, no gene flow (= speciation complete)

![]()
If no gene flow after secondary contact, speciation was completed in allopatry. Speciation would then be viewed as a byproduct of divergence in allopatry. What happens after secondary contact is a matter of great debate: If the two differentiated forms mix or hybridize this may provide the context for selection for assortative mating also called reinforcement of premating isolation (reinforcement hypothesis). In this case speciation was not completed in allopatry and fate of the two populations depends on the outcome of the interaction upon secondary contact.
Patterns predicted from the action of reinforcement:

![]()
Selection in zone of overlap for increased premating isolation. See artificial demonstration with a selection experiment (fig. 16.4, pg. 432).
Reinforcement model assumes that hybrids are less fit
(=means by which selection for further isolation can operate). This assumes
that post mating barriers arise first and that premating barriers
arise as a result of selection in sympatry; these assumptions may
not hold in all cases. However, if premating barriers evolved first, there
might be little hybridization (speciation complete?); if there was no postmating
barrier, even with small amounts of hybridization the two forms would fuse
back together because there would be no selection against hybrids!
Reinforcement is actually a special case of character displacement
which is the accentuation of differences between species (or forms) by
selection against the individuals of similar phenotype (reinforcement =
reproductive character displacement and is achieved by selection against
hybrids). If reinforcement is true, we should expect to see displacement
of characters associated with pre-mating barriers to gene exchange in areas
of secondary contact. Some cases we do: calling songs of anurans;
Frequently such reproductive character displacement is not observed.
When "reinforcement-like" patterns are observed, one has to be
sure that the phenotypic shift is actually an evolutionary response to
the presence of the other incipient species and not to some other clinal
variation (e.g., ecological factors that generate parallel clines).
Problems with reinforcement: other possible outcomes: fusion of the two populations because differentiation was sufficiently slight that selection against hybrids is weak relative to the gene flow between forms. extinction of one or the other of the two forms. Quite likely when there is selection against heterozygotes. In population genetic terms, equivalent to heterozygote disadvantage AA, Aa, aa with fitnesses 1,1-s,1, a metastable equilibrium
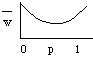
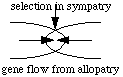
Selection against hybrids within the zone of secondary contact only
favors displacement in sympatry; gene flow in from allopatry will swamp
the effect. One could view such hybrid zones as genetic canyons
of lost alleles. Another important question: if selection against hybrids
is the driving force for reproductive character displacement, how will
the genes for the different components of isolation/recognition sweep
through the allopatric regions of the two species ranges where there
is no hybridization, hence no selection?
Another allopatric model is the Peripatric model referring to populations surrounding the main part of the current species range. See fig. 16.5, pg 434.
1. Small isolated populations
2. Genetic drift via population bottleneck or founder event => new allele frequencies
3. new "genetic environment" => different response to selection than in main population
4. effect is a major genetic change = "genetic revolution"
![]()
![]()
One consequence is that speciation may not be dichotomous. Important
consequence: rapid divergence, unlikely to leave fossil intermediates
(these possibilities will come in to play when we discuss "punctuated
equilibrium" later).
A variant on this theme proposed by Hampton Carson an influential evolutionary biologist from the University of Hawaii is Founder-Flush speciation :
1. population initiated with small number of individuals (founders)
2. flush in population size; relaxed selection during this phase; low fitness recombinants survive
3. crash in population size; selection and drift determine which genotypes survive.
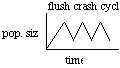
Carson's view: two "parts" to the genome: the "open
variability system" and the "closed variability system"
Open system has much variability, responds rapidly to selection
(loci encoding allozyme polymorphisms such as enzymes in glycolysis and
Krebs cycle, etc.); closed system is resistant to selection;
less variable ) loci encoding courtship song, developmental patterns,
etc.) In Carson's view the closed system is reorganized during
the flush-crash cycles, leads to a genetic change that contributes to reproductive
isolation/mate recognition.
Questions about the founder flush speciation: how small is population after crash?, how long does population stay at reduced population size? Could retain a large portion of the genetic variation after one crash; extended bottle necks will be more effective in reducing variation.
These questions also could apply to Mayr's peripatric speciation model
Parapatric Model of speciation. Ranges of two differentiated forms are contiguous and non-overlapping. Patterns of discontinuities between differentiated forms/populations may be due to secondary contact after a period in allopatry, or the discontinuity could be due to primary differentiation in situ. One cause of this might be a steep environmental gradient or habitat boundary (see fig. 16.3 pg. 428). With selection on loci that affect reproductive isolation/mate recognition, populations can become differentiated. Will be apparent in the formation of a cline. Can lead to sufficient divergence of reproductive/mating characteristics that barrier to gene flow is established (e.g., plants growing on mine tailings have diverged in flowering time).

Studies of parapatric distributions are frequently concerned with the
concordance of clines. Selection acting on one locus/trait can impose
a cline on another character if the two characters/loci are linked.
Are clines superimposed, shifted, different slopes. Slatkin (1973)
has shown that the width of a cline is: See
fig. 16.9, page 439; text uses different letters for equation). Different
loci may have different cline shapes due to different strengths of selection
acting on them.
The text is a bit misleading about Parapatric speciation. It might lead
one to believe that when a hybrid zone is observed, parapatric speciation
is involved. This is not true since the hybrid zone may be the result of
secondary contact after allopatry, rather than primary differentiation
at the hybrid zone interface. Here again we need to determine the relative
importance of the allopatric phase and the parapatric interaction in determining
the outcome of speciation (or fusion). The cricket hybrid zone (fig. 16.8,
page 438) is in fact the result of allopatry followed by secondary contact
(my personal knowledge), but Ridley does not let you know this.
Non-allopatric models of speciation are controversial but not impossible. Sympatric speciation can be modeled with a two locus polymorphism, one locus (A) affecting fitness (in this case by affecting fitness in terms of survival on one of two alternative hosts/patches), and another locus (B) affecting mate choice which is crucial in the evolution of assortative mating, a barrier to gene exchange (proposed by John Maynard-Smith in 1966)
| AA | Aa | aa | BB | Bb | bb | ||
| host 1 | 1+s | 1 | 1 | mate w/ AA | no preference | mate w/ aa | |
| host 2 | 1 | 1 | 1+t |
These selective regimes maintain polymorphism at the A locus as in a multiple niche polymorphism considered in the population genetics section. These sets of fitness/mating values will result in the evolution of associations (e.g., linkage disequilibria) between the A and the B locus (e.g., AABB individuals and aabb individuals will be found in the populations with few intermediates.
![]()
![]()
these have high fitness these have low fitness
The green lacewings (Genus Chrysoperla; formerly Chrysopa)
seem to exhibit patterns of host preference and mate choice similar to
that presented above (studied by the Taubers, Cornell University). One
form is adapted to one host/habitat and a second to another; this habitat
preference appears to be controlled by a single locus with other modifying
loci (some evolutionists have not accepted the lacewing data as conclusive).
See the other sympatric speciation model that involves variation in
a resource base (fig. 16.10, pg. 443). This model still requires the evolution
of associations (e.g., linkage disequilibrium) between fitness genes and
behavior genes.
But, if sympatric speciation is, if not common, at least possible, is
the model really sympatric?: is it just microallopatric speciation
(some argue NO if adults come up off their hosts into a mating swarm, but
then proceed to mate). Another crucial issue is: what is the rate of
recombination between these two types of loci since crossing over will
break up favorable associations. A model of host preference and assortative
mating invoking many genes (polygenic model) make it more difficult
to maintain nonrandom associations. A general issue with all of these models
is how much gene flow is tolerated. Evolution of barriers to
gene exchange is the issue, gene flow = gene exchange; how much gene
flow can take place and still evolve barriers to the gene flow??
The answer depends on the genetic architecture of speciation (how
many genes, how much divergence, etc.; next lecture on genetics of speciation).
Saltatory speciation: Richard Goldschmitt in the Material Basis of
Evolution proposed the idea that Macromutations (mutations with
big effects) would result in major developmental and phenotypic changes
in their carriers producing the so-called hopeful monster. Ridiculed
at the time; recently gained a new readership due to the molecular characterization
of genes that cause major phenotypic effects (more later on evolution of
development). Big problem remains: who is the hopeful monster going to
mate with?
Chromosomal speciation: Consider a diploid with 2N = 4 chromosomes.
If two such individuals failed to undergo the reduction division of meiosis
their gametes would be 2N=4. If these gametes were used in fertilization
of one another, a new chromosomal number would be established: 4N = 8.
If this became stabilized as a new chromosomal type (and this is common
in plants), this new type can be reproductively isolated from the original
2N = 4 species. The reproductive isolation would be due to an imbalance
of chromosome sets in the new zygote: N = 2 gamete crossed to an N = 4
gamete results chromosomal type of 3N = 6. There can be two consequences
with this imbalance: i) inviability due to failure during development or
ii) instability during chromosome segregation could result in gametes with
an incomplete set of chromosomes (aneuploidy). These consequences could
have the effect of a reproductive barrier between the original 2N
= 4 and the polyploid 4N = 8 type. Speciation can be nearly instantaneous
when such chromosomal events are involved (multiples of even numbered ploidy
levels: can produce gametes with some exceptions; multiples of odd numbered
ploidy levels: usually cannot produce gametes due to imbalance of haploid
complements) => speciation. Thus polyploid hybrids are frequency
genetically isolated from their progenitors.
The simple inversion model (figure 16.17, pg. 457) illustrates another
way that chromosomal factors might play a role in speciation.
How should we think about speciation events? What are the models of
divergence: is speciation like a peak shift in an adaptive landscape, or
is speciation a gradual divergence process on a flat adaptive landscape?
Main issue is whether the peak itself shifts and hence the population
shifts with it, or whether the two alternative peaks already exist
and the problem is shifting between the two alternatives.
Some fundamental issues in thinking about speciation: 1) does speciation require allopatry or can speciation occur in non-allopatric contexts (sympatric, parapatric)?; 2) does speciation require changes in many genes or can changes in a few specific genes lead to speciation?; 3) is speciation itself adaptive or does speciation occur as a byproduct of adaptive responses to other pressures?; 4) what determines the rates of speciation? (some lineages speciate at very different rates).